Day 2: Using secondary antibodies!
If the primary antibody which was used the day before came from a mouse, then a goat anti-mouse secondary antibody needed to be used ie. the antibody was created in a goat and upon being exposed to mouse antibodies, it synthesized specific antibodies against it.
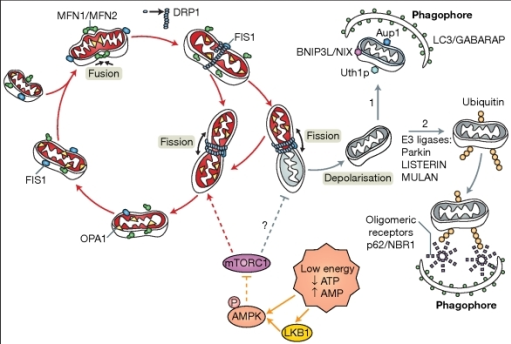
After the membranes were treated with the secondary antibodies, they needed to be visualised using LiCOR, a software which is a special imaging system than allows us to visualise the fluorescence of the secondary antibodies and thus the presence or absence of our desired protein, Parkin (52kDa).
As well as visualising Parkin, we needed to also standardise our data with controls by locating ubiquitous proteins GAPDH and TOMM20.
GAPDH (glyceraldehyde 3-phosphate dehydrogenase) is a 37kDa protein which is found in the cytosol of all cells and takes part in the 6th step of glycolysis during the process of anaerobic ATP production.
TOMM20 is a 29kDa mitochondrial membrane protein which is found on the surface of all mitochondrial membranes and is responsible for the translocation of proteins that were synthesised in the cytosol, into the mitochondria.
The results
Despite the fact that this memory was a bit messy, I was happy with my very first result. Eventhough the antibodies unfortunately did not work very well for Parkin, there were clear bands for GAPDH at 37kDa in the cytosolic contalateral and ipsilateral sample lanes. This result was expected seeing as GAPDH is a cytosolic protein.
Again, Parkin was unfortunately not visable on this membrane, but I was happy with the membrane appearance and the clear presence of TOMM20 at the mitochondrial contralateral and ipsilateral sample lanes. This result was also expected as TOMM20 is only a mitochondrial protein.






Comments
Post a Comment