Day 8: Transfecting the C.17 cells
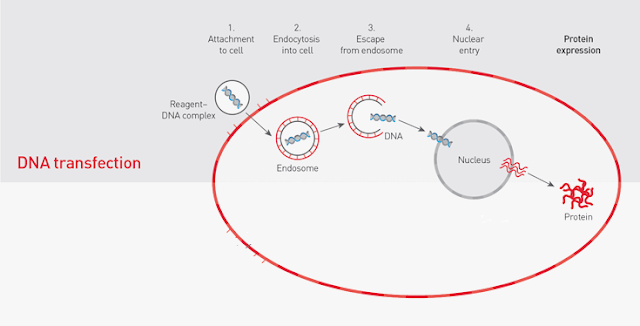
We decided to transfect the C.17 cells with lipofectamine 2000, which is a lipid rich mixture containing lipid micelles that a capable of transporting genetic information extra cellularly to the intracellular environment. The lipid micelles contain hydrophilic heads and hydrophobic tails that can fuse with the C. 17 cells.
What is Keima?

Keima is a special coral derived acid stable fluorescent protein which can fluoresce different colours depending on the pH that it is exposed to. The Keima cDNA is ligated into a plasmid with a mitochondrial tag at its N terminus. The DNA that we wanted to transfect was a plasmid called Keima.
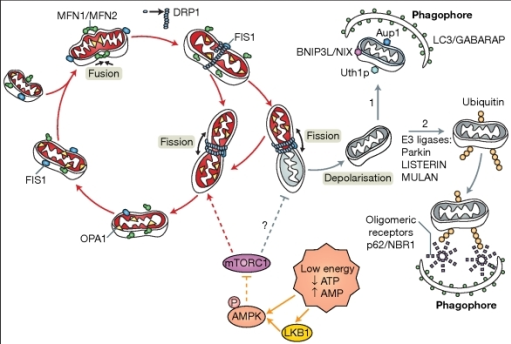
In order to understand how Keima works, it is important to get an insight on how mitochondrial mitophagy works
The mechanism of mitophagy (Exner et. al., 2012)
We decided to transfect the C.17 cells with lipofectamine 2000, which is a lipid rich mixture containing lipid micelles that a capable of transporting genetic information extra cellularly to the intracellular environment. The lipid micelles contain hydrophilic heads and hydrophobic tails that can fuse with the C. 17 cells.
The liposome has a cationic exterior and is able to attract the overall partial negative charge on the cDNA of the plasmid. Once inside, the liposome releases the plasmid which is taken up into the nucleus, where it is transcribed into its corresponding mRNA transcript. After post translational modification, the mRNA transcript is translated to the protein product: Keima.
What is Keima?

Keima is a special coral derived acid stable fluorescent protein which can fluoresce different colours depending on the pH that it is exposed to. The Keima cDNA is ligated into a plasmid with a mitochondrial tag at its N terminus. The DNA that we wanted to transfect was a plasmid called Keima.
In order to understand how Keima works, it is important to get an insight on how mitochondrial mitophagy works
The mechanism of mitophagy (Exner et. al., 2012)



Comments
Post a Comment